Microbial Pathogenesis Unit of Navarrabiomed-UPNA identifies new gene structure in bacteria that may trigger novel developments in the fields of synthetic biology and bacterial biotechnology
The Microbial Pathogenesis Unit of Navarrabiomed-Public University of Navarra (UPNA) has found a new genetic organisation in bacteria that helps better understand bacterial biology. The study of this genetic architecture was published in Proceedings of the National Academy of Sciences of the United States of America (PNAS).
Research background
In 1961, François Jacob and Jacques Monod discovered that bacteria group the genes that encode the proteins for a certain metabolic pathway in a single transcription unit (which they called ‘operon’). They won the Nobel Prize in Physiology or Medicine in 1965 for their discovery.
The bacteria they chose for their study was Escherichia coli, which normally lives in the intestines of healthy people; specifically, they studied the set of genes E. coli bacteria need to transport lactose (milk sugar) and break it down. E. coli only produces the three proteins it needs to digest lactose when the sugar is available. To simplify transcription regulation, the three genes involved are adjacent in the genome and under a single regulation system. Similar transcriptional regulation systems are found in other metabolic pathways in all bacteria.
Research at Navarrabiomed
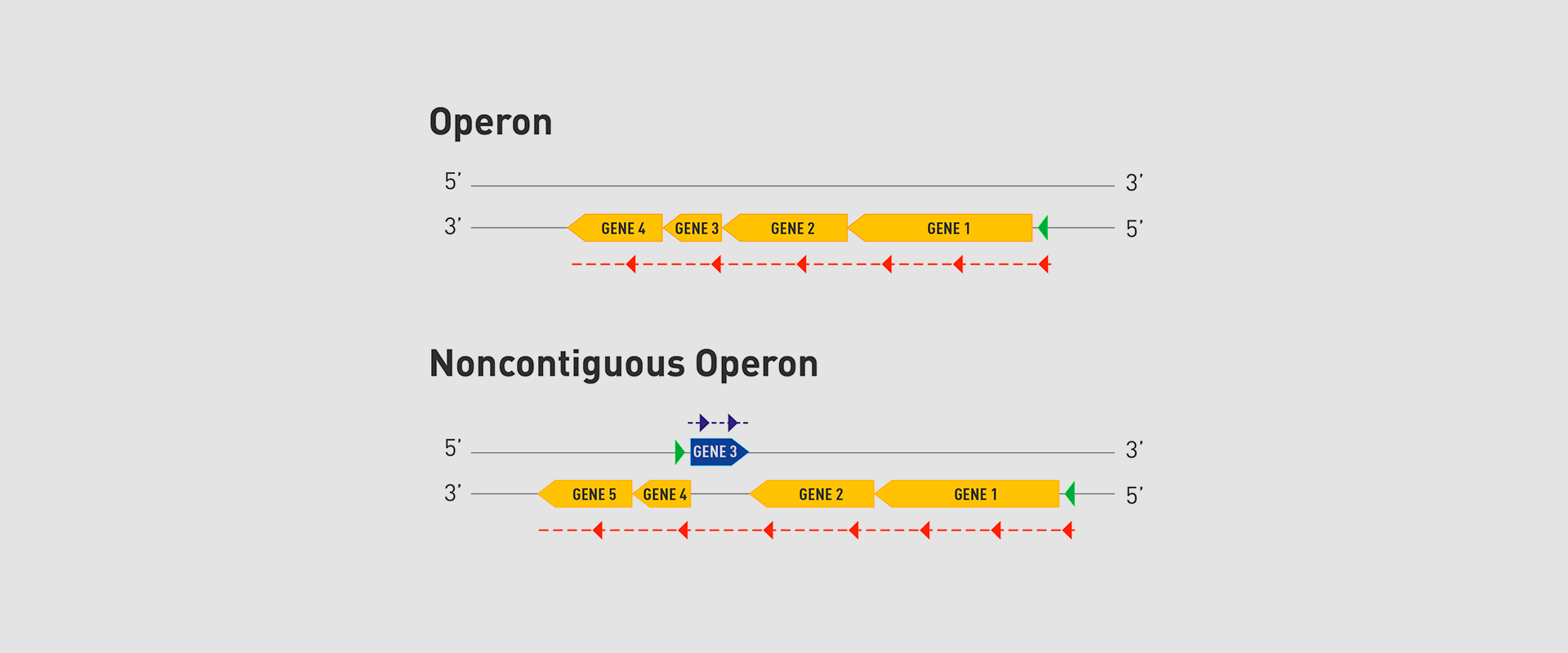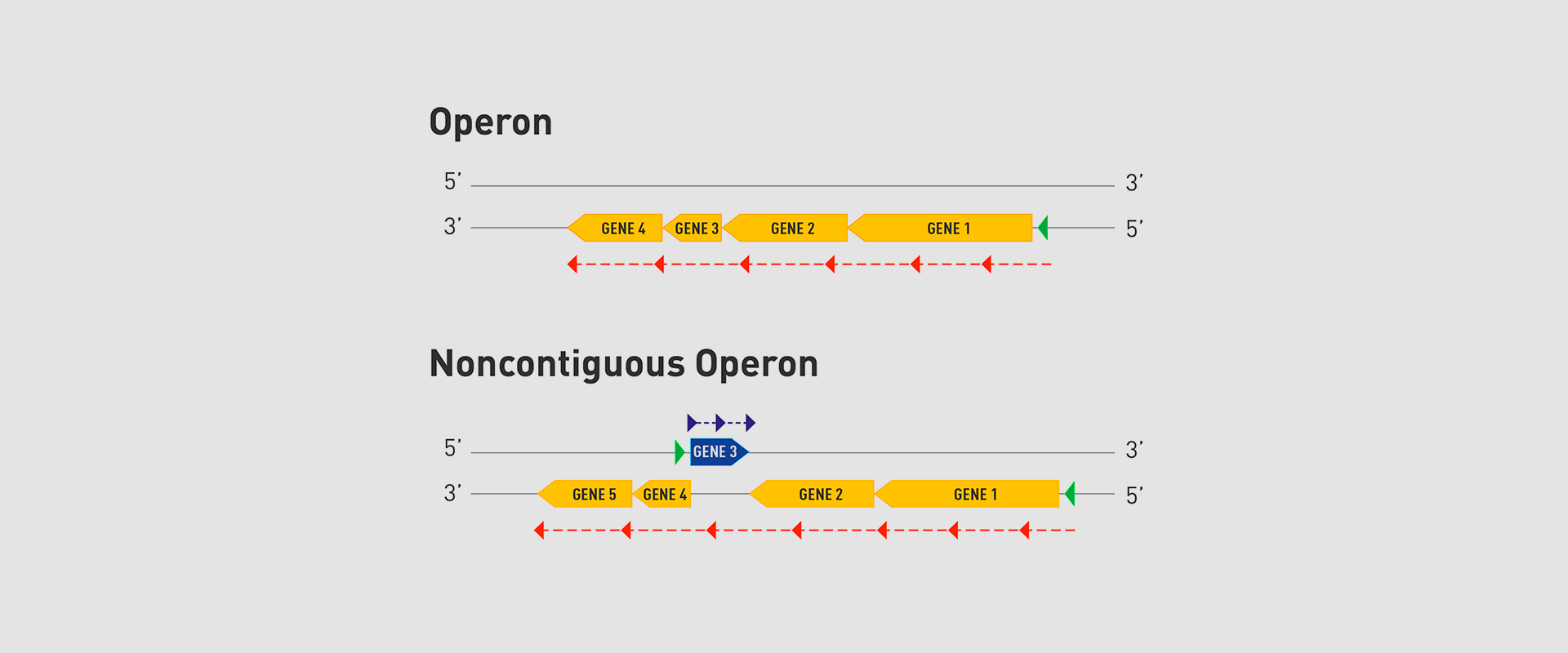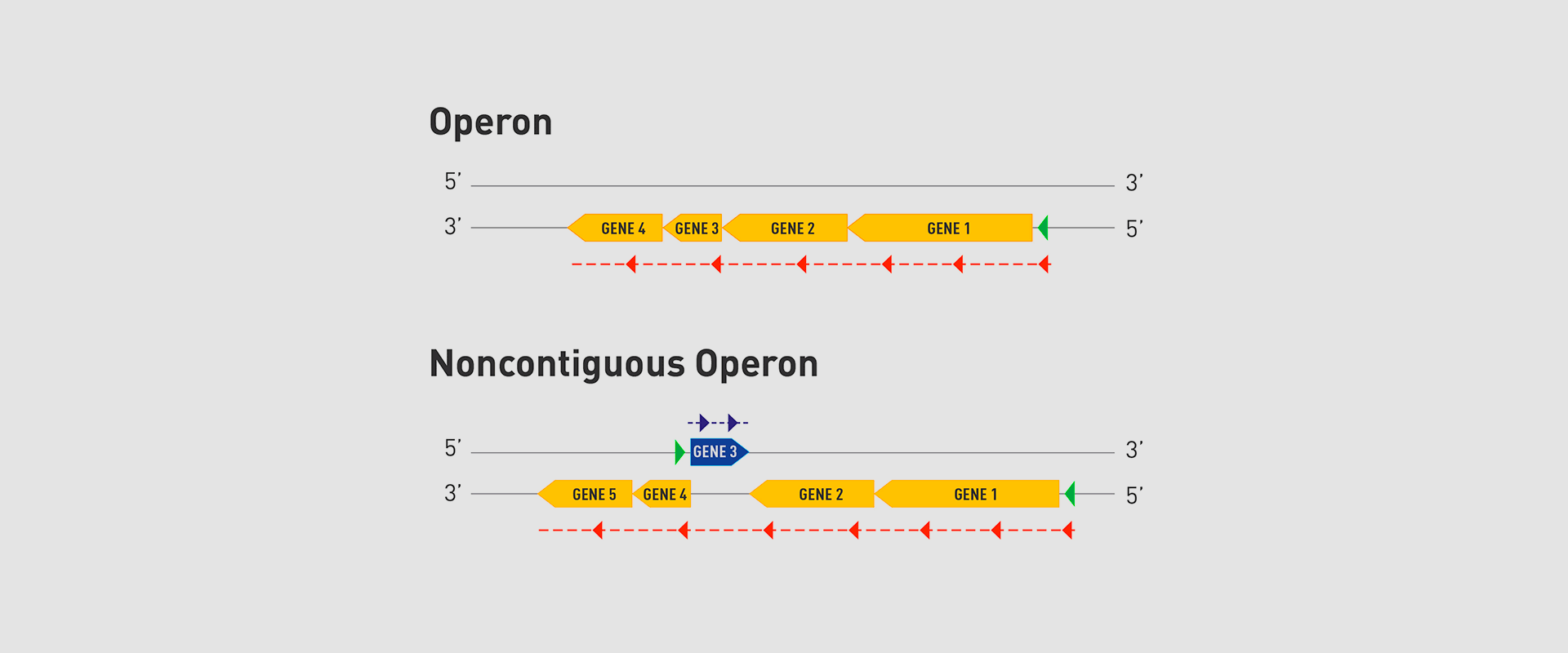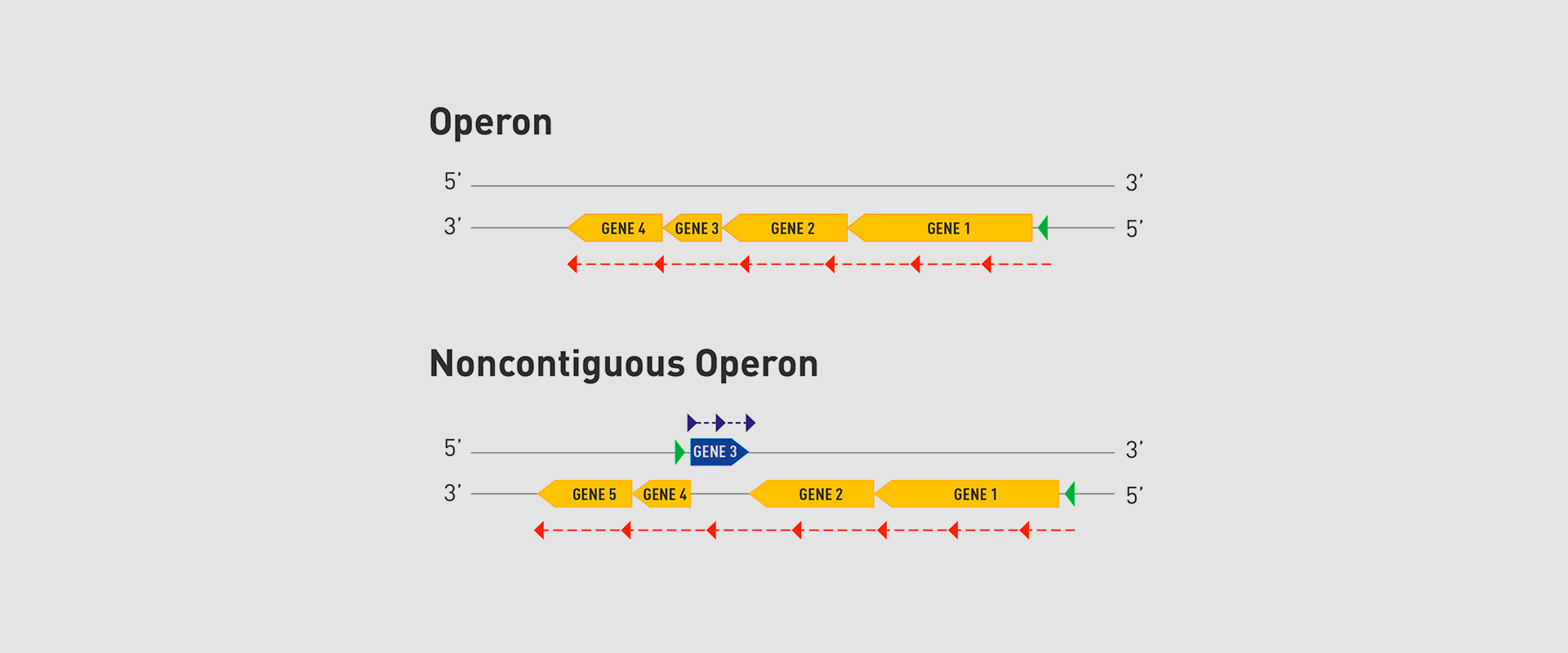
In 2018, the team of researchers at Navarrabiomed coordinated by Iñigo Lasa Uzcudun, Head of the Microbial Pathogenesis Unit and Director of the biomedical research centre, described a new way genes are organised in bacteria. This regulation system has a higher level of regulation in operon structure, which the authors of the study named ‘non-contiguous operon’.
The bacterial model analysed has a group of four genes that are transcribed as a transcription unit despite the existence of a separate gene between the second and third genes that is transcribed in the opposite direction.
This transcriptional architecture results in an antisense transcript that acts as a mutual regulation system for the expression of the genes in the operon and the gene that produces this antisense transcript. Therefore, the concept of non-contiguous operon includes not only the genes transcribed from the same transcription unit but also overlapping genes whose expression is coordinated with that of the genes in the operon.
This finding deepens the understanding of bacterial biology and may trigger novel developments in the fields of synthetic biology and bacterial biotechnology.
The study was carried out as part of the scientific activity done at the Navarra Medical Research Institute (IdiSNA).
